CSD-Discovery Workshops
These self-guided workshops are tutorials intended to demonstrate the various tools and features of the CSD-Discovery suite including GOLD, CSD- CrossMiner, Hermes.
Tell us what you think!
We hope you found these workshops helpful. As we aim to continuously improve our training materials, we would like to hear your feedback. You can do so by filling out this survey. When asked, insert the workshop code that you find in the workshop description in this page. It will only take 5 minutes and your feedback is anonymous. Thank you!
Introduction to Full Interaction Maps
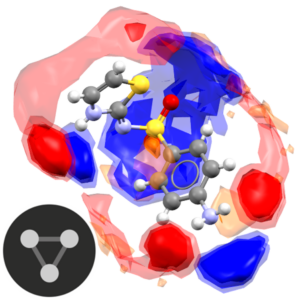
This is a self-guided workshop on how to use the full interaction maps for exploring crystal structures; including assessing hydrogen bonding and substituent effects.
Prerequisites: This handout is also part of the CSDU module Analysing intermolecular interactions 101 – Full Interaction Maps. Visit the module webpage to learn more about FIMs, and for more demos and tips.
Workshop code: MER-002
Understand Protein-Ligand Interaction Patterns Using Full Interaction Maps (FIMs)
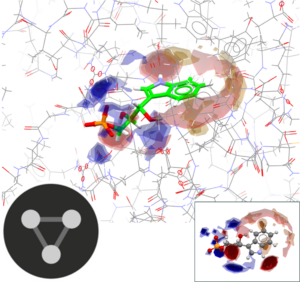
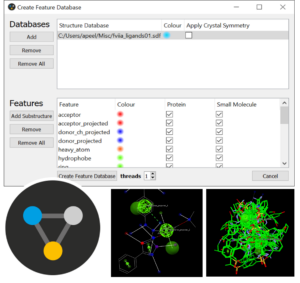
This is a self-guided workshop on how to generate full interaction maps of protein-ligand interaction patterns and analyse docking solutions with FIMs.
Prerequisites: Introduction to Hermes (HERM-001)
Workshop code: MER-004
How to Generate Ensembles of Conformers using the CSD Conformer Generator
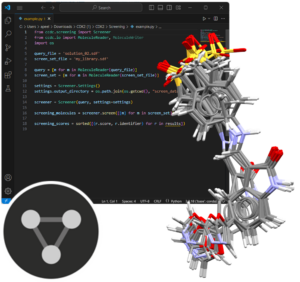
This is a self-guided workshop on how to generate ensembles of conformers using the command line function, Mercury and the CSD Python API
Prerequisites: Basic knowledge of Mercury, if you are new to Mercury, you can start with the Visualisation workshop – MER-001, try the CSDU module Visualization 101 – Visualizing structural chemistry data with Mercury. A basic understanding of Python and the command line, Introduction to the CSD Python API – PYAPI-001.
Workshop code: CONF-002
Introduction to Protein-Ligand Docking with GOLD
This is a self-guided workshop on the basics of protein-ligand docking in GOLD, demonstrating how to set up, run and analyse your docking results.
Prerequisites: Introduction to Hermes – HERM-001
Workshop code: GLD-001
Ensemble Docking with GOLD
This is a self-guided workshop on the how to perform non-native docking of a ligand into an ensemble of conformers, accounting for movements of the protein backbone.
Prerequisites: Introduction to Hermes – HERM-001, Introduction to Protein-Ligand docking in GOLD – GLD-001
Workshop code: GLD-002
Pose Prediction with GOLD
This is a self-guided workshop on the how to perform pose prediction with GOLD, focusing on building a correct representation of the protein binding site and choosing high accuracy settings.
Prerequisites: Introduction to Hermes – HERM-001, Introduction to Protein-Ligand docking in GOLD – GLD-001
Workshop code: GLD-003
Pharmacophore Constraints in GOLD
This is a self-guided workshop on the how to include pharmacophore constraints during protein-ligand docking in GOLD.
Prerequisites: Introduction to Hermes – HERM-001, Introduction to Protein-Ligand docking in GOLD – GLD-001
Workshop code: GLD-004
Flexibility and Constraints in GOLD
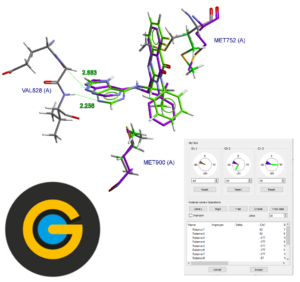
This is a self-guided workshop on the how to inspect a protein crystal structure to determine how to include flexible sidechains in your sampling.
Prerequisites: Introduction to Hermes – HERM-001, Introduction to Protein-Ligand docking in GOLD – GLD-001
Workshop code: GLD-005
Docking with Waters in GOLD
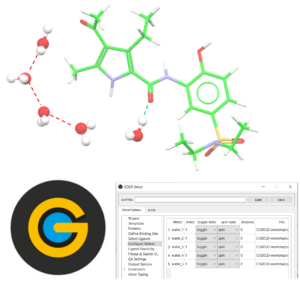
This is a self-guided workshop on the how to perform docking into a binding site that contains water molecules.
Prerequisites: Introduction to Hermes – HERM-001, Introduction to Protein-Ligand docking in GOLD – GLD-001
Workshop code: GLD-006
Structure-Based Virtual Screening with GOLD
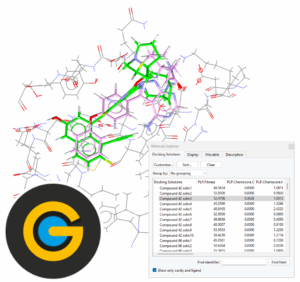
This is a self-guided workshop on how to conduct a structure-based virtual screening using GOLD.
Prerequisites: Introduction to Hermes – HERM-001, Introduction to Protein-Ligand docking in GOLD – GLD-001
Workshop code: GLD-007