Short Guide to CIFs
A CIF or Crystallographic Information File is the standard format for storing crystallographic structural data. CIF information has a specific structure or format that needs to be followed to allow crystallographic programs to read the file.
CIFs are usually created automatically by the program used to process or refine crystallographic data; this means it is very unlikely that you will need to create a CIF from scratch (and we don’t recommend editing CIFs by hand). However, being able to read and find information within a CIF is useful and can help you ensure you provide complete crystallographic information for your structures, as well as interrogate the data of others.
Learning Outcomes
This guide will introduce you to the type of information that can be included within the CIF, as well as give more details information about the CIF format and how information is stored within a CIF.
It aims to:
- Increase your familiarity with the CIF format.
- Help you to become more comfortable when reading a file, checking and correcting it, as well as when extracting information.
What is in a CIF?
What kind of information can I find in the CIF?
A CIF contains information about the crystal structure (such as unit cell values, atom names and their coordinates and any structural model quality indicators, e.g., R Factor) as well as any details of the diffraction experiment (such as temperature, pressure, experimental wavelength and the type and name of equipment used) and any data processing undertaken (such as the programs used to process the data).
The material that is included within a CIF varies and depends on what information the authors of the structure incorporated, as well as the technique and program used. Many crystallographic programs include most information automatically when the CIF is made. However, some items may not be included in the CIF as they are not relevant to a particular experiment or may be missed by the program (if they are included in log files created by other programs that were not kept in the final refinement folder or if the refinement package being used does not check them when compiling the final CIF).
Most CIFs deposited to the CCDC also contain reflection intensity data (or .hkl information). This is the data that the crystal structure model is refined against, which comes from the processing of the experimental diffraction data or images.
When producing a CIF, it is important to include as much information about your structure in your CIF to ensure your data is reproducible.
Viewing a CIF
The CIF format was specifically designed to be readable by both humans and machines. Many crystallographic programs can read and extract information from a CIF. CIFs can also be read by humans in many text editors.
The CCDC has a free CIF reader and editor called enCIFer, which has some handy additional features such as colour coding, syntax checking and the ability to visualise the structural information in 3D. enCIFer can also identify syntax violations and contains two data entry wizards: one for experimental data about your structure and the other for publication details, to help you ensure you’ve provided as much information as possible about your structure. enCIFer syntax checking is also embedded into the CCDCs online deposition service enabling you to check and correct the format of your CIFs as well as enhance your datasets during the deposition process.
Further information about enCIFer, including a user guide and tutorials, can be found here.
enCIFer Interface - Viewing the CIF

enCIFer Interface - Visualizing the Structure

CIF Format
The CIF contains the crystallographic information for a structure in a well-defined and standardised format. We will describe it in the following section.
Starting a CIF
The information for one structure/experiment is called a data block. A CIF can contain multiple data blocks.
A data block begins with the string ‘data_’ to signify the start of the structural information. This may be followed by a block code. The block code is a name string for the data to help identify the structure. No spaces are allowed in the block code. Within a CIF, each block code should be unique and no two data blocks can have the same block code.
In Example 1, ‘lcysteine’ is the block code of the data block. The block code is case-insensitive.

Example 1: The start of a data block: data_lcysteine (colour coded in red)
In enCIFer, the ‘data_’ string and the block code is colour coded red.
CIF Fields
Within a data block, each piece of information is stored in a CIF field. A CIF field is made up of a data name, which describes a piece of information about the structure, and a data item, the information related to the data name.
Data names are used to identify the information and begin with an underscore ‘_’ followed by the name of the field with no whitespace. A data name must only appear once in a particular data block. Data names are case-insensitive: in a CIF _cell_volume and _Cell_Volume are equivalent data names.
In Example 2: ‘_cell_volume’ is a data name while 526.44(4) is the data item.

Example 2: A data block with a CIF field: data_lcysteine (colour code in red) (new line) _cell_volume (colour coded in blue) 526.44(4)
In enCIFer, data names are colour coded blue.
The relevant CIF fields to store data can be found in CIF dictionaries. The IUCr maintains dictionaries of standard data names to store information for different techniques (https://www.iucr.org/resources/cif/dictionaries). The dictionaries contain a list of data names and a definition of the allowed data item response for each data name, including any required scientific units.
Storing Data
Data items are stored in the CIF in a variety of ways. The relevant CIF dictionary should be consulted for specific guidelines for each field (including any required units to store data in).
If the information for the data item is unknown, or the CIF field is not applicable to the experiment, then a placeholder item can be used instead: ‘?’ is used for unknown data, while ‘.’ is used for inapplicable CIF field.
1. Single value data items
Data items which are a single value (with no spaces) are reported after the data name with whitespace between the data name and the data item.

Example 3: Single value data items
2. Short data items
A data item that is longer than a single value or input (but does not run over multiple lines) is reported by providing the information within matching ‘ ’ single or “ ” double quotation marks.

Example 4: Storage of short data items
3. Longer data items
Longer data items spanning multiple lines are enclosed by semi-colons ‘;’, which illustrate the beginning and end of the response. The semi-colons must be at the start of the line to be recognised as enclosing the response. In enCIFer information enclosed within semi-colons will be a dark green colour.
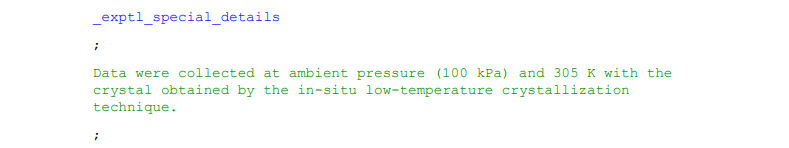
Example 5: An example of data spanning multiple lines
NOTE: One common CIF syntax error is the incorrect semi-colon usage. If a semi-colon is missing or out of place in the CIF, the CIF may be incorrectly read, and important data not recognized by any program. In these instances, in enCIFer, you may notice that the colour coding in the CIF file is not as expected.
4. Loops
Data items can also be stored as part of a loop. This is usually used to store a table of values, such as atom coordinates. The loop begins with ‘ loop_’ (colour coded in pink in enCIFer), which is then followed on subsequent lines by the data names in the table. After that, the value data is stored: each line will contain a data item for each of the data names listed. Each value is separated by a white space.
Loops can also contain placeholder items for any unknown or not applicable data items – these need to be included otherwise the information in the loop will not be read properly. An example of this is the value for U iso of the atom labelled H1 in Example 6.
Only one level of loop is allowed, which means that there can be no nested loops inside each other.
Example 6: Atomic position data stored in a loop
Other Format Information
Comments: Any line beginning with a ‘#’ is classed as a comment in a CIF and not read by crystallographic programs. Lines that are comments in CIFs appear in a dark green colour in enCIFer.
# This is a comment
White space: The information in CIF fields does not have to be aligned with other values, although this may make it easier for humans to read the file. Both tabs and spaces count as white space.
Allowed characters: The allowed characters within a CIF are space, horizontal tab, new line , carriage return and the following characters:
‘ ! ” # $ % & \’ ( ) * + , – . / 0 1 2 3 4 5 6 7 8 9 : ; < = > ? @ A B C D E F G H I J K L M N O P Q R S T U V W X Y Z [ \\ ] ^ _ ` a b c d e f g h i j k l m n o p q r s t u v w x y z { | } ~
Unknown data items or not applicable CIF fields: In cases where the information for the data name is unknown, the data item for that data name should be given as ‘?’. If a CIF field is not applicable to the experiment, the data item should be given as ‘.’.
Case sensitivity: Data names and data block codes are case-insensitive. The case of any data items should be respected by CIF reading software.
Storing Additional Data
How do other files get embedded in the CIF?
Most modern crystallographic refinement programs embed the model files in the CIF file by default. By including the ‘ACTA’ command in your refinement instructions file (.ins), a CIF will automatically be created and embed the .res (updated refinement instruction file after refinement) and .hkl files.
Older versions of refinement programs (e.g., SHELXL-97) may not do this. It is recommended to refine your structure using up-to-date software (e.g., SHELXL 2014 or later) if possible.
How do I Know if my CIF is Correct?
enCIFer can alert you to any problems with the syntax of your CIF. The feedback is provided on the bottom left hand of the window in the form of Errors, Warnings and Remarks (see image 1) . Errors are serious problems with your CIF syntax. Warnings are less serious syntax errors, data names that aren’t currently found within the IUCr CIF dictionaries or where data values do not conform to the allowed values (as specified in the CIF dictionaries). Remarks indicate that mandatory data items are missing from the CIF. Further information about Errors, Warnings and Remarks in enCIFer can be found in the enCIFer User Manual .
The IUCr’s checkCIF service also includes a CIF Syntax check. This web service allows a user to check their CIFs for problems with their structural model, missing or inconsistent data, as well as highlight potential issues with the model quality. It creates a checkCIF report, which provides feedback as a series of Alerts (A, B, C and G in order of seriousness) that the user should aim to resolve before publication if possible.
When depositing a CIF to the CCDC using the joint CCDC and FIZ Karlsruhe web deposition service (for depositing data into the CSD and the ICSD) there is also a syntax check stage. Any syntax errors will be highlighted and must be fixed before the deposition process is allowed to continue. This online deposition process also incorporates the IUCr’s checkCIF service.
How to Edit CIFs
Some programs, including enCIFer and our online deposition process, have methods of helping you add information about your structure without having to manually edit the CIF yourself. enCIFer contains two data entry wizards, one for publication details and the other for chemical, physical and crystallographic properties, which can help you supplement the data in your CIF. These wizards can be found in the Tools Menu within enCIFer, as ‘Publication Wizard’ and ‘Crystal Wizard’ respectively.
It’s Your Turn
Can you answer these questions about CIFs based on what you’ve read in this short guide?
- How many times can a CIF data name appear in a data block?
- What information should be stored in the _cell_volume CIF field? Hint: try looking for the field name in the IUCr CIF dictionary.
- What is the x coordinate for atom N1 in Example 6?
- What character is used to indicate that a CIF field is not applicable to a particular experiment?
Additional Resources
CIF Format
- More detailed descriptions of the CIF format
- The IUCr CIF dictionaries
- IUCr guide to CIF for authors
- Hall, S. R., Allen, F. H., & Brown, I. D. (1991). The Crystallographic Information File (CIF): A new standard archive file for Crystallography. Acta Crystallographica Section A Foundations of Crystallography, 47(6), 655-685. doi:10.1107/s010876739101067x
- Educational material on CIF available from the Database of Educational Crystallographic Online Resources (DECOR).
enCIFer
CIF Checking
- IUCr’s CheckCIF service
- Further information about checkCIF checks
- Spek, A. L. (2020). Checkcif validation alerts: What they mean and how to respond. Acta Crystallographica Section E Crystallographic Communications, 76(1), 1-11. doi:10.1107/s2056989019016244