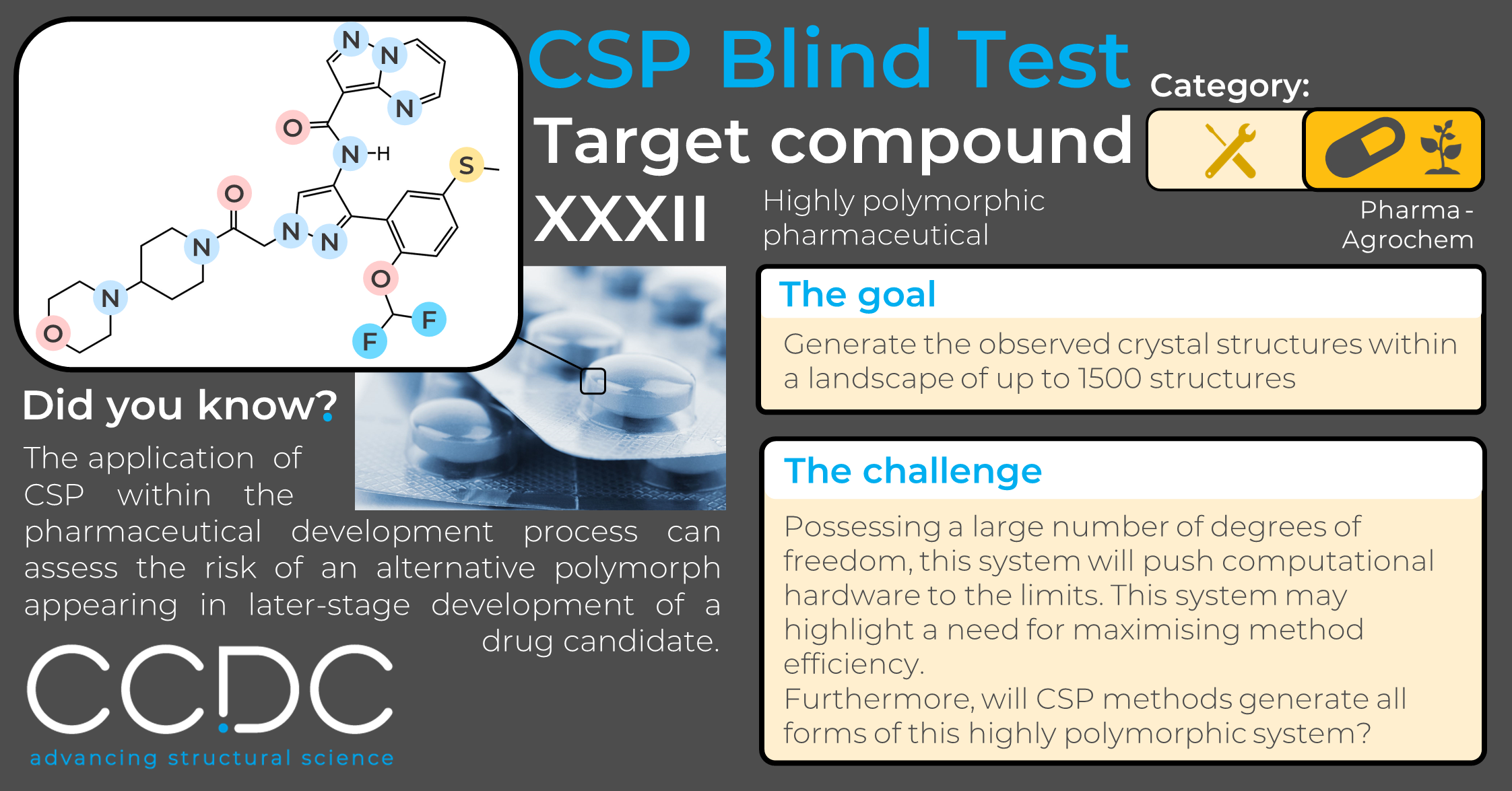
CSP Blind Test Structure Reveal – Target XXXII
Here, we reveal the 2D chemical structure of one of the most challenging systems included in CSP Blind Test history. Excitingly, it is much more representative of the complex-natured pharmaceutical compounds that are commonly encountered in the present day.
Revealing the target compound structures
For the first time since the initial CSP Blind Test in 1999, we are releasing the 2D chemical structures of the target compounds to the public through our website and social media platforms. We hope that this will showcase to the wider community the types of systems that CSP methods are applicable to, but more importantly, expose the challenges that CSP currently faces in terms of both computational hardware limitations (due to molecular size) and chemical complexity, and encourage wider participation for finding suitable solutions
The structure
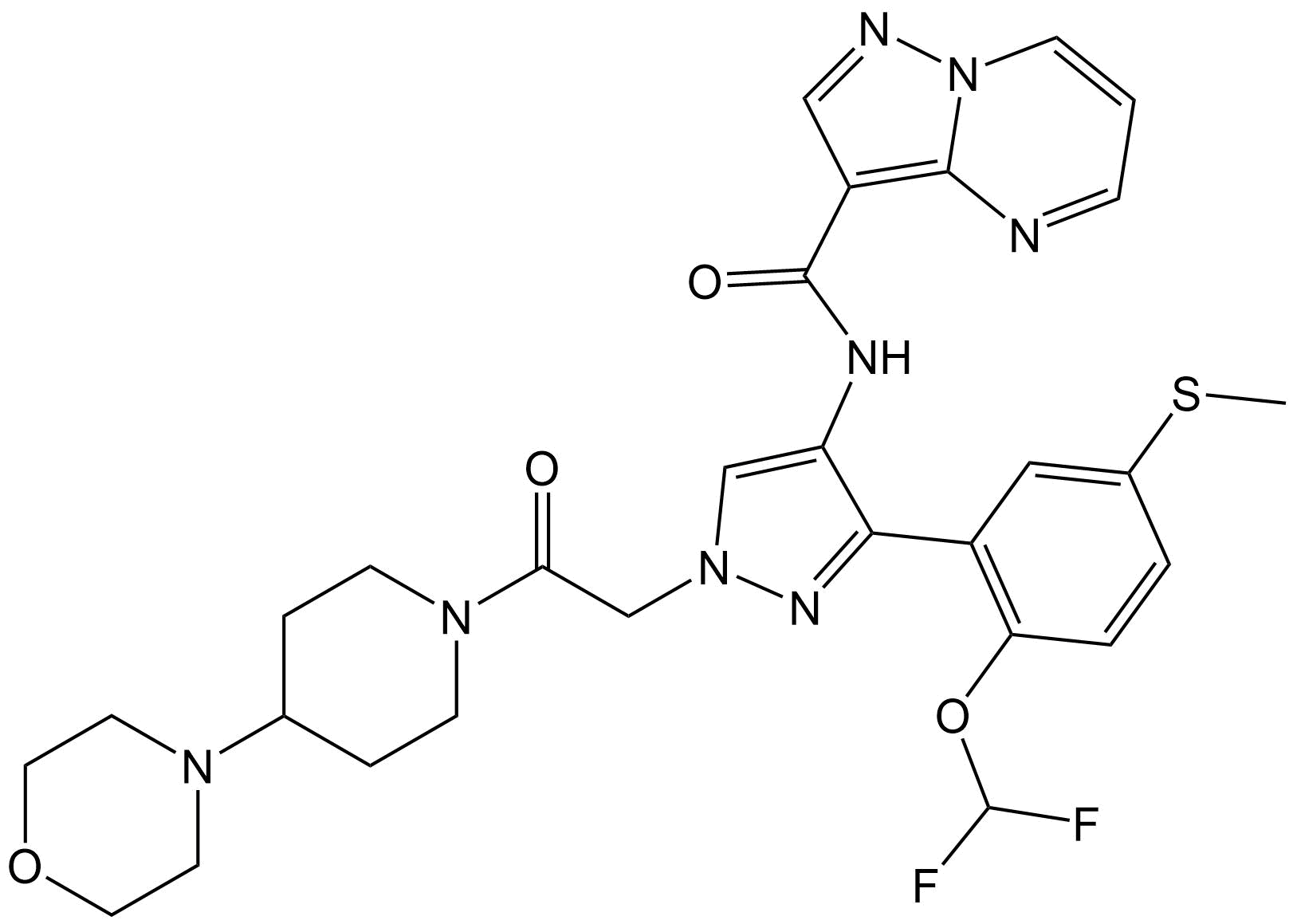
Target XXXII is a large, highly polymorphic, pharmaceutical drug candidate.
The application of CSP to pharmaceutical systems
CSP can be used to analyse the risk of a late-appearing polymorphic form at later-stage development of pharmaceutical drugs by identifying alternative polymorphs that may be thermodynamically more stable. This situation can be disastrous, as was the case for Ritonavir, a life-saving treatment for AIDs introduced to the drug market in 1996.[1] In 1998, a new polymorphic form appeared which showed dramatically less solubility and bioavailability, meaning production stopped and complete reformulation was required for the drug. With the rapid development of CSP methods since this time, pharmaceutical companies are more informed of these types of risks, and can make appropriate decisions earlier within the development process.
The challenge
Included within the ‘Pharma-agrochem’ category, participants are asked to submit a landscape of up to 1500 predicted structures in which the CCDC will analyse whether the experimentally observed form(s) are predicted.
Participants will also be given a prepared list of 500 structures, one or more of which represent the experimentally observed form(s), and asked to rank the list using their own method in order of relative stability. The CCDC will analyse whether the experimentally observed form(s) are ranked within the most stable of the submitted list.
How does this test CSP methods?
Having the greatest number of rotatable bonds, this system will be pushing computational hardware to the limits, and with a whopping 8 known polymorphic forms, the big question will be whether all forms are generated by CSP methods. This is debatably the most complex system included in the history of the CSP Blind Test, and excitingly it is much more representative of the complex-natured pharmaceutical compounds that are commonly encountered in the present day.
References
[1] Bauer, John, et al. “Ritonavir: an extraordinary example of conformational polymorphism.” Pharmaceutical research 18.6 (2001): 859-866.