How Your Structure Is Curated in the CSD – The Journey of a CSD Entry
The Cambridge Structural Database (CSD) is the world’s largest database of curated small molecule organic and metal-organic crystal structures. Each single entry is curated by our scientific editors to maintain the standards of the database, making the CSD a valuable and trusted source of experimental crystallographic data.
In this blog, we will follow the journey of a new CSD entry, and dive deeper into the process of data curation, from enhancing the data that we receive, to making it more accessible and discoverable.
The CSD Data at a Glance
The CSD contains a stunning total of 1,329,543 unique curated crystal structures from the contributions of 512,675 different authors worldwide (data updated as of February 18th 2025, additional details can be found at CSD Statistics).
The database contains crystallographic data of organic and metal-organic compounds, which can be further categorized as drug molecules, agrochemicals, pigments, explosives, protein ligands, coordination polymers, and metal-organic frameworks. Where the data is available, the CSD also captures additional information such as:
- Melting points
- Crystal colours
- Crystal habits
- Bioactivity details
- Natural source data
- Oxidation states
- Polymorph families
Every year, between 50,000 and 60,000 new structures are made available to the public through the CSD following our data curation processes.
The following sections will show the journey of a structure, from deposition through to publication, and underpin each step of the data curation process.
Data Deposition
The journey of a structure starts with the deposition of its CIF in the CSD, which is primarily done by the authors or crystallographers, and less often sent by the publishers. You can deposit your structure via our online deposition service by signing in with your CCDC account.
A variety of structural information can be added when an author deposits a structure to enrich the CSD entry. This includes entering a response for any major alert generated from the automatic and optional IUCr checkCIF service, details about the associated publication, data related to the experimental determination, and physical properties of the crystal.
At this stage, the author can also decide to share the structural data directly in the CSD, without having any associated scientific publication. For more details on this, visit our CSD Communications page.
The deposited data is stored privately until the associated article is published. However, journal editors and peer-reviewers can access the data before its publication to support their evaluation.
Once the CIF is deposited, we have a workflow in place with all the major publishers to allow us to update the publication details automatically and have the correct structural data assigned to the corresponding article.
Data Publication
When the article associated with the new structure is published in the literature, the structural data is made available to the public on WebCSD, our online searching tool.
Up until this stage, only automated checks have been performed on the deposited data. Hence, a message that explains to the users at what stage of curation the entry is will appear from the information button on the top of the WebCSD visualizer (Figure 1).
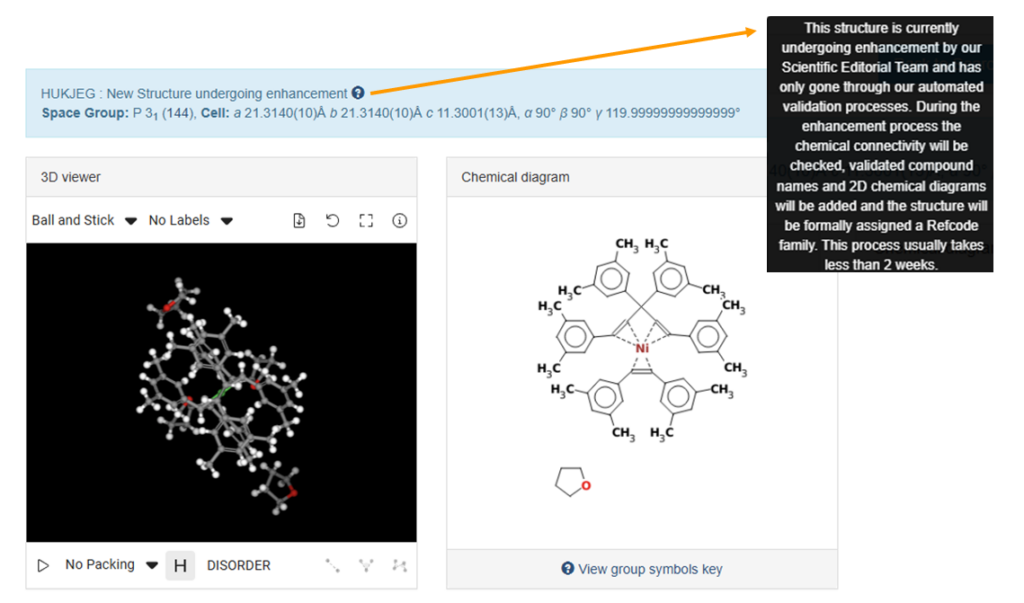
Deposition Numbers, CSD Refcodes, and Data Credit
On deposition in the CSD, the structure is automatically given a Deposition Number or CCDC Number. Upon publication, the structure becomes associated with a CSD Refcode, a unique 6-8 character identifier.
To make the CSD entries sharable, citable, and attributable, a Digital Object Identifier (DOI) is assigned to each of them.
At the CCDC, we believe that the collective use of data impacts the scientific progress and that each entry can make the difference. For this reason, we make your data accessible and reusable for others in the community, for today and for the future, while advocating for recognition and credit to be given to the authors. This is one way we adhere to the FAIR Data Principles, which ensure the data in the CSD is Findable, Accessible, Interoperable, and Reusable.
Why Do We Curate Your Experimental Data?
You might be wondering: now that the original data is published, why do we provide further curation? Are there any benefits?
Firstly, at the CCDC we prioritize quality. The CSD is a trusted resource, being relied upon by industry and academia. Therefore, it is vital that we perform checks on quality and accuracy of the deposited data.
Secondly, with over 1.3 million structures, it is important we maintain consistency, readability, and discoverability of the data. What we aim to do is to annotate and enhance the data with metadata such as names, diagrams, and properties. All these aspects contribute to making sure that the wealth of data is accessible and discoverable to our users.
Finally, as well as being a wealth of knowledge to researchers, helping to inform and progress their work, the CSD offers several software suites which provide additional insights using the underlying CSD data, making curation even more important.
Automated Data Curation
As mentioned previously, automated checks and data processing are performed upon deposition of a structure in the CSD. These include:
- Duplicate checks
- Syntax checks
- CCDC Number assignment
- CIF information extraction
- Bond assignment
- Disorder resolution
- 2D diagram generation
- Naming
- CSD Refcode assignment
The outcome of this automated process is what is accessible immediately after publication on WebCSD.
Automation speeds up the process of curation, allowing each of our editors to process up to 100 entries per day (compared to 20 per day prior to automation). It is generally reliable for small organic molecules, which usually need small intervention. However, it can’t be a complete replacement of human input.
Manual Data Curation: The Human Touch
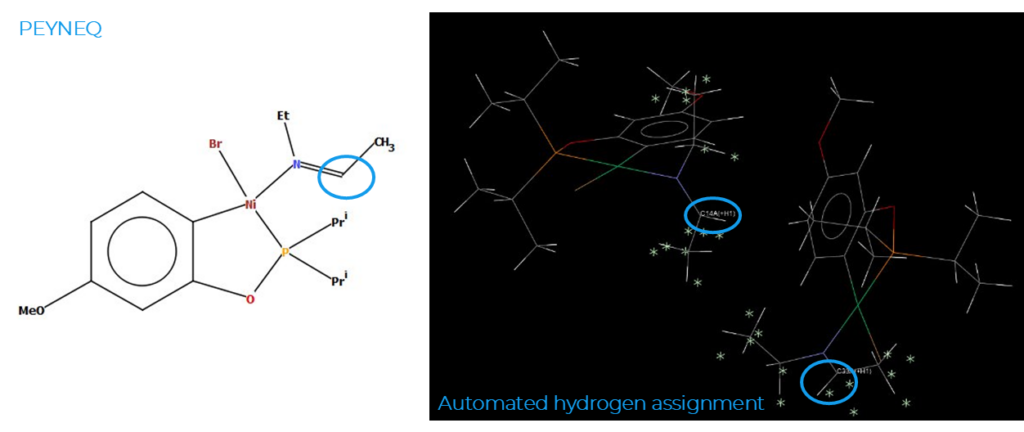
The manual data curation and enhancements done by our editorial team start with checks of the chemical connectivity in the structure to make sure that the charge is balanced, that the valency, hydrogen assignment, and stoichiometry are correct, and that they match the author description.
In the example shown in Figure 2, it can be seen that the automated bond assignment performed upon deposition for CSD Entry PEYNEQ (Figure 2, right) didn’t match the chemical structure reported in the paper (Figure 2, left). More specifically, in the automated step an extra hydrogen atom was added to the carbon atoms highlighted by the blue circles (Figure 2, right), which was then removed during the manual curation step to mirror the correct bond assignments.
This represents a case in which the solely automatic hydrogen assignation would have given the wrong hybridization to a carbon atom in the structure, and hence would have been chemically incorrect.
Another aspect that requires manual checks and intervention is disorder. To start with, our editors populate or edit the disorder field with general or more descriptive comments on the type of disorder present in the structure. The population of this field raises the disorder flag that allows disordered entries to be filtered out when searching the CSD.
Previously, when non-symmetry based disorder was present in a structure, this was often manually resolved by suppressing the lower occupied sites. For symmetry based disorder, instead, the molecule was fully suppressed for clarity only if it was a solvent, while any other portion of a structure was kept as it was to make it clear to the users that a disordered component was present in the 3D coordinates. We now have the capability to add disordered atoms in disorder assemblies and groups to allow users to get a better appreciation of disordered geometries. More information can be found in our blog ‘Handling Disordered Structures in Mercury and the CSD Python API’.
Our team also manually adds any additional information about disorder and void space, if present, including the use of SQUEEZE or MASK.
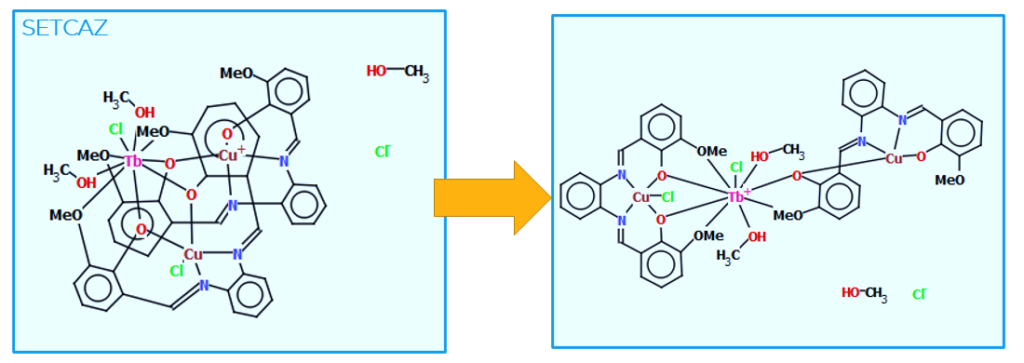
Following the disorder treatment, the manual curation continues with edits of the 2D chemical diagrams to make them readable, clear, avoid overlaps, and to maintain the CSD consistency. Importantly, these diagrams allow structure-based searching, and hence they need to be further checked to make sure they are faithful to the authors description.
From Figure 3 it can be seen that initially the 2D diagram of CSD Entry SETCAZ is very unclear. After undergoing manual curation, users can clearly visualize the two organic ligands in the structure and get a better understanding of how the coordination spheres of the metal centres are populated.
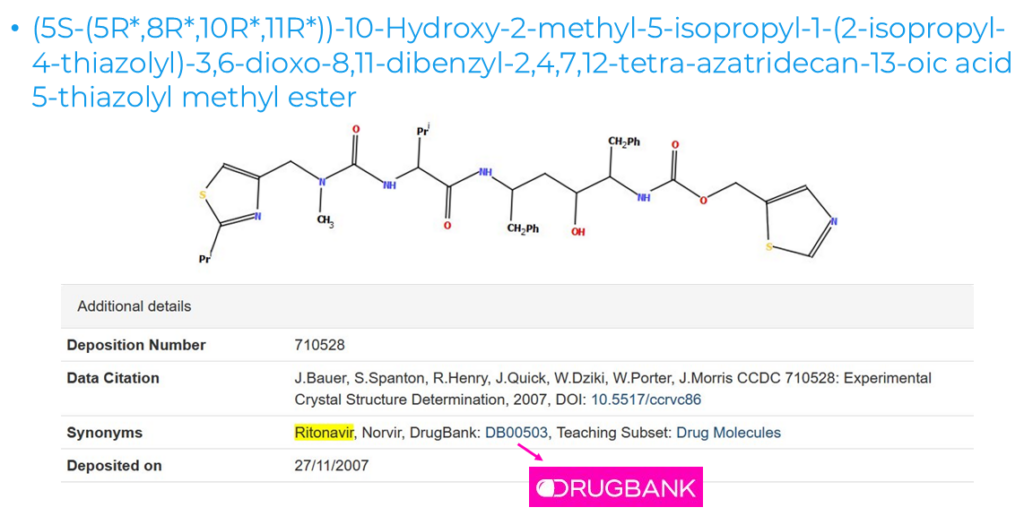
The following step in the manual curation is the generation of the IUPAC standard compound names, including edits to make them readable and findable, and addition of extra information, such as oxidation states, stereochemistry, links to external databases, and synonyms for drugs or materials such as MOFs.
We can see an example of this in Figure 4, where the synonyms Ritonavir and Norvir were manually added to the details listed for CSD Entry YIGPIO. Users can hence find the structural details of this well-known drug very easily by using its common name instead of the more complex IUPAC name. The drug can also be searched by its DrugBank number.
Our editors can also further enhance the CSD entries by adding additional information about bioactivity, melting point, pressure, and phase transitions. They also assign of a Refcode family, if applicable, for polymorphic structures or for data collections of the same crystal structure determined by different research groups, at different pressures or temperatures, for example.
When Is the Curated Data Made Available?
Around 95% of the data we receive are curated within a month of publication. However, some structures require extra curation time to ensure the underlying data is correct and/or an accurate representation is made available. This includes addressing severe disorder, manually naming complicated structures, and in some cases contacting the author for further clarification.
It should be noted that while we enhance the deposited data, we do not change the underlying data, which is available to download as the original CIFs from WebCSD.
Only when a structure meets all the quality standards of the CSD, its full curation process is considered completed.
Conclusions
In this blog, we followed the journey of a new CSD entry and we highlighted the enhancements that each entry receives during our data curation processes.
The CSD is a valuable and trusted source of experimental crystallographic data for many scientists worldwide. Our scientific editors will continue to curate the entries in the database to maintain its standards and to ensure that the data is accessible and discoverable.
Nest Steps
- Discover the different research areas the Cambridge Structural Database (CSD) can support.
- To discuss further and/or request a demo with one of our scientists, please contact us via this form or .