InChIs, Stereochemistry, and ChemDraw Export – New Interoperability Initiatives
The new release of our desktop software includes interoperability initiatives including InChIs in the CSD, stereochemistry search, and ChemDraw export.
InChIs in the CSD
Support for the IUPAC International Chemical Identifier (InChI) in our database, software and workflows is now embedded, enabling the reliable lookup of structures and interoperability between CCDC and external systems.
As the InChI has attained the status of an industry standard, researchers can readily benefit from synergies across a diverse range of scientific resources. Embedding support for the InChI in our database, software and workflows enables easy and reliable retrieval of data from the CSD using a standard identifier and easy intersection of CSD data sets with other information resources using a standard identifier.
InChIs in the CSD allows users to read the ‘stereochemical layer’ of InChI strings, one factor that now allows our stereochemistry search to be improved.
Stereochemistry Search
Currently our tools can read and display stereochemistry, but search options have been limited.
Stereoisomers are hard to distinguish using other common search criteria in the CSD. Using stereochemistry as a search constraint will help find relevant structures. Recognizing stereochemistry from input files will save time and improve accuracy when constructing search queries, increasingly important as the CSD continues to grow beyond its 1.2M structures.
To begin to address this, our search code that can be accessed through the CSD Python API has been enhanced allowing users to find search results that match the stereochemistry constraints that interest them.
The new functionality allows users to:
- Set stereochemical restraints
- Conduct substructure searches taking account of the set stereochemical constraints
- Conduct substructure searches to recognize enantiomers in a crystal.
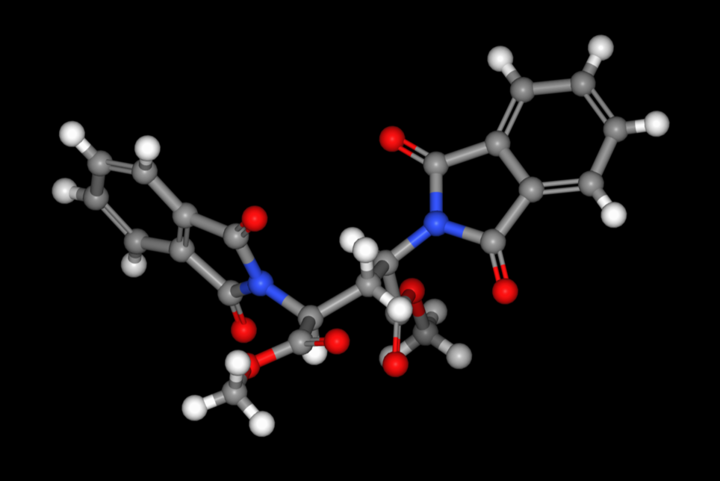
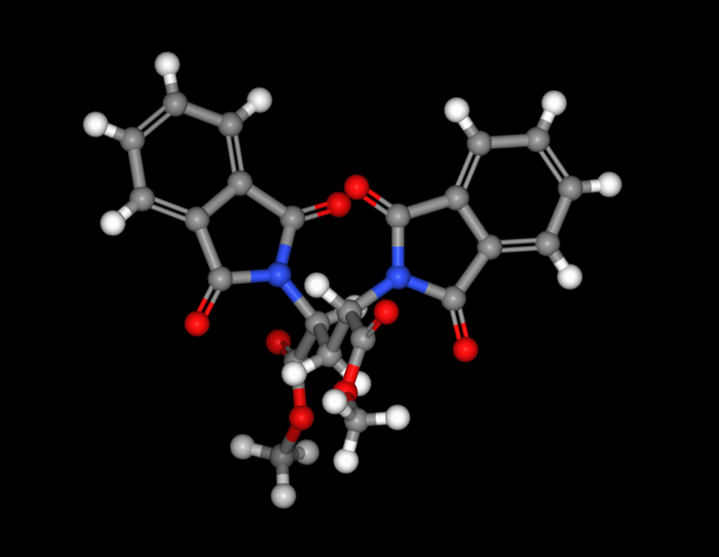
In addition, the stereochemical input from 2D and 3D input files is now also recognized when searching the CSD.
The SMILES to Molecule tool has also been released in Mercury. SMILES (Simplified Molecular Input Line Entry Specification) are a way to represent a molecule in a compact way, readable by both machines and humans. They are a standard language used in chemistry and cheminformatics to represent molecular structures.
ChemDraw Export
You can now import a CCDC diagram into ChemDraw, allowing further clean up and improvements. simply save your viewed chemical diagram as ChemDraw XML and then Load the saved file into ChemDraw. This will save time when writing presentations and reports with industry-standard diagram styles.
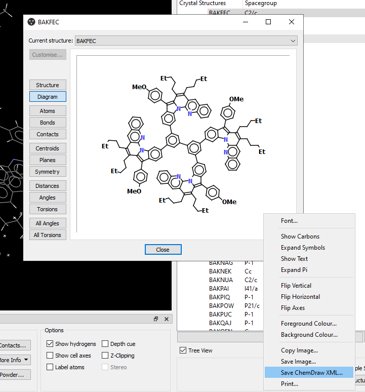
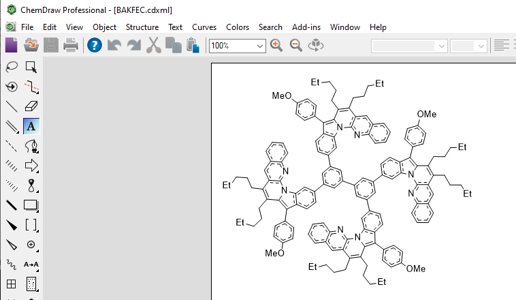
Learn More
Try out these new features for yourself in:
- Mercury – view, Analyse, and Understand Molecular Structures and Their Properties
- Hermes – view, Analyse, and Explore Macromolecular Structures and Their Properties
- CSD Python API – programmatically Access Structural Chemistry Data in the Cambridge Structural Database and Perform Custom, Repeatable Molecular Analyses.
- The Cambridge Structural Database – the Comprehensive Repository of Validated and Curated Small Molecule Organic and Metal-organic Crystal Structures.